Annotation of animal genomes: genes, promoters, functional motifs, protein sub-cellular localization.
Softberry software to analyze animal genome sequences:
- Ab initio gene prediction: Fgenesh (3180 citation of using in genomic projects according to Google scholar) with total 506 genome-specific parameters for various organisms, including 329 parameters for animals.
- Gene prediction using similar protein support: Fgenesh+
- Automatic Genome Annotation pipeline Fgenesh++ (see flowchart) including known genes (mRNA) mapping (Est_map) and database protein mapping (Prot_map) to apply Fhenesh+
- Fgenesh++est Anntotaion pipeline improving predictions using mapped ESTs (include real non-coding non-coding 5'-and 3'-gene ends, when possible)
- TSSG/W, FPROM - Promoter prediction for human genes
- Nsite - search for functional motifs in promoter sequences
- ProtComp - prediction of protein localization in cellular compartmen
- Software for training parameters of gene-prediction programs for new genomes
Evaluations:
NGASP Gene prediction competition: http://www.wormbase.org/wiki/index.php/Results
The nGASP project parallels recent computational prediction initiatives including CASP, GASP, and EGASP. A summary of the results will be submitted for peer-reviewed publication. For nGASP, a set of regions representing ~10% (10 Mb) of the C. elegans genome release WS160 was selected to evaluate the performance of the participating gene predictors.
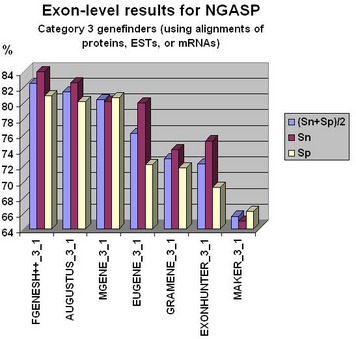
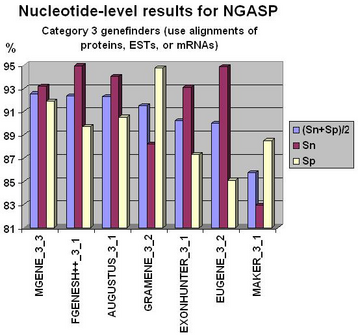
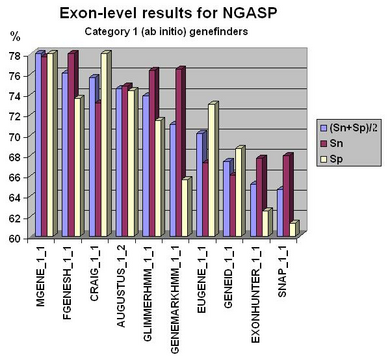
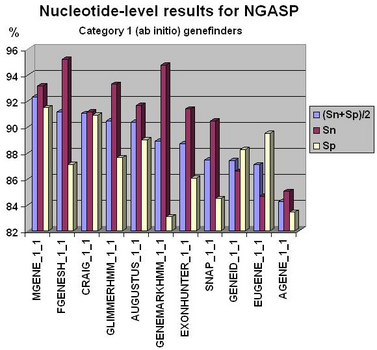
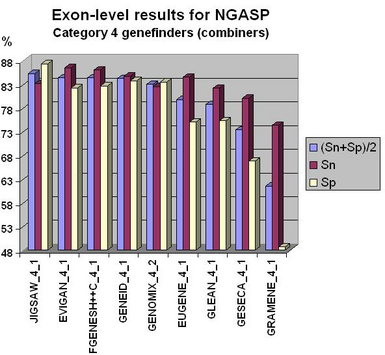
Fgenesh++ pipeline was the best on exon level gene prediction in recent NGASP Gene prediction competition. In most other categories Fgenesh and Fgenesh++ have the best sensitivity (with Specificity close to the best). We give the preference to sensitivity as any genomic region in tests can have some undiscovered genes. Even in the combiner category we have comparable results with the best combiners, while we do not use other predictors results, just a variant of fgenesh++ with improved EST accounting (see following histograms).
 |
 |
 |
 |
 |
Application for animal genome analysis in selected publications:
Fgenesh group software: Nature (2008) 453, 1064 - 1071; Nature (2008) 452, 949 - 955; Nature (2008) 452, 88 - 92; Nature (2008) 451, 193 - 196; Nature Biotechnology (2008) 26, 553 - 560; Nature Genetics (2007) 39, 715-720; Nature Biotechnology (2007) 25, 319-326; Molecular Microbiology (2007) 64, 3, 755-770; Nature (2006) 443, 931-949; PNAS (2006) 103, 43, 15794-15799; Nucleic Acids Research (2006) 34, 17, 4685-4701; Nucleic Acids Research (2005) 33, D399-D402; PNAS (2005), 102, 5, 1566-1571; Nature Genetics (2004) 36, 40-45; Nature Biotechnology (2004) 22, 1146-1149; PNAS (2003) 100, 11, 6569-6574. Google Scholar shows ~500 research publications (Keywords: fgenesh and prediction) using fgenesh software for gene finding in many genomes).
ProtComp: PLoS ONE (2008) 3(6): e2300; Planta (2008) 227, 491-503; Current Genetics (2008) 53, 217-224; Nature Protocols (2007) 2, 953-971; Nature (2006) 444, 97-101; Genes & Development (2006) 20:1365-1377; Journal of Lipid Research (2006) 47, 268-283; Microbiology (2006) 152, 547-554; Genome Research (2003) 13, 2265-2270; PNAS (2001) 98(9): 5341-5346.
Nsite: Eukaryotic Cell (2008) 7, 6, 988-1000; DNA and Cell Biology (2008) 27(6), 307-314; European Journal of Human Genetics (2007) 15, 463 - 472; Molecular Microbiology (2007) 66, 2, 534-551; Insect Science (2007) 14, 1, 5-14; Epilepsy Research (2007) 75, 2-3, 145-153; Mammalian Genome (2006) 17, 8, 892-901.
Fgenesh, TSSW/G, Nsite and ProtComp have also been used in patents and patent applications.