The application provides the possibility to apply different color schemes to the symbols of residues. Selecting a color scheme can be performed using the "Color" group's commands of the main menu. The coloring may occur either for the symbol of residue or for its background (it depends on settings defined in the "Options" dialog box, see "Options dialog").
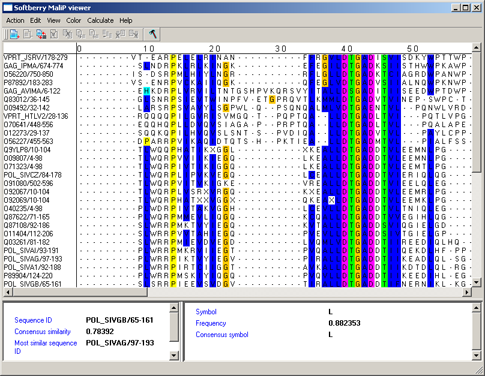
ClustalX colors - ClustalX colors - The color scheme used in the "ClustalX" application [2]. The color of a symbol depends on residue type and on frequency of its occurrence in the column (see fig. 1, tab. 1).
 |
Figure 1. |
Table 1.
| Residue type | Conservatism | Color |
| Hydrophobic (ACFHILMVWY) | >60% | BLUE |
| With negative charge (DE) | >50% | MAGENTA |
| With positive charge (KR) | >60% | RED |
| Polar (STQN) | >50% | GREEN |
| Cysteines | >85% | PINK |
| Glycines | >85% | ORANGE |
| Prolines | >85% | YELLOW |
| Aromatic (FYW) | >50% | hydrophobic CYAN |
Zappo colorscheme - aminoacidic residues are colored in accordance to their physicochemical properties (see fig.2, table 2).
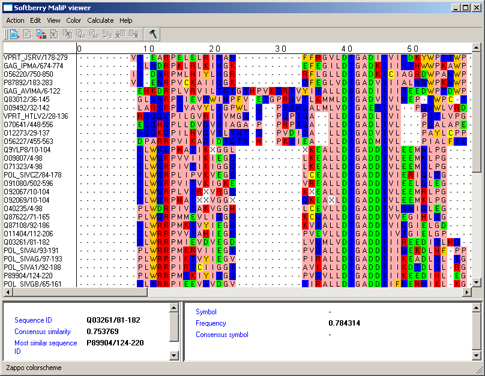 |
Figure 2. |
Table 2.
| Residues | Description | Color |
| ILVAM | Aliphatic/Hydrophobic | Pink |
| FWY | Aromatic | Orange |
| KRH | With positive charge | Red |
| DE | With negative charge | Green |
| STNQ | Hydrophilic | Mid blue |
| PG | Proline/Glycine (conformationnaly special) | Magenta |
| C | Cysteine | Yellow |
Taylor colorscheme - Taylor colorscheme - aminoacidic residues are colored in accordance to the scheme provided by Taylor [3] (fig. 3).
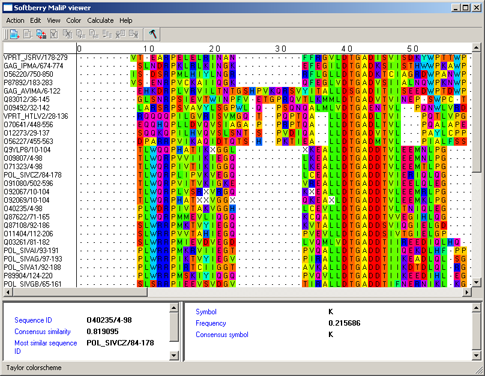 |
Figure 3. |
By hydrophobicity - aminoacidic residues are colored in accordance to the table of hydrophobicity by [4]. According to the table the most hydrophobic residues are colored in red, the most hydrophilic ones - in blue. Colors of residues with intermediate properties are the halftones of purple, which depend on the scale value (fig. 4).
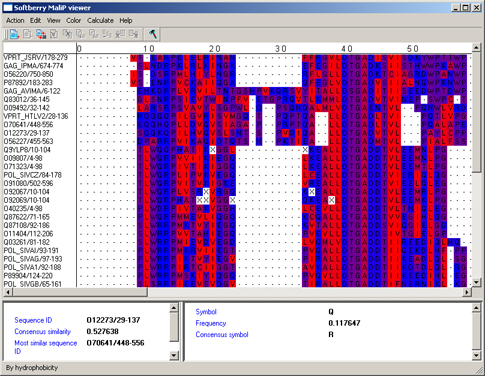 |
Figure 4. |
Helix propensity - preferring formation of the a-helix (fig. 5) [5].
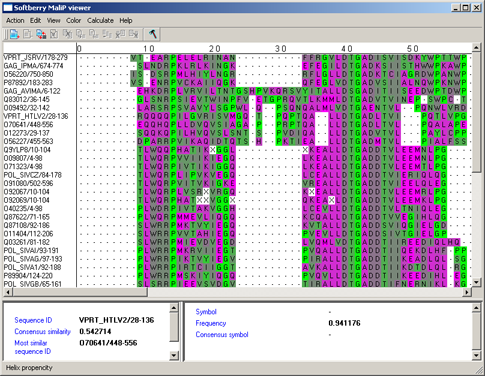 |
Figure 5. |
Strand propencity - preferring formation of the b-folds (fig. 6) [5].
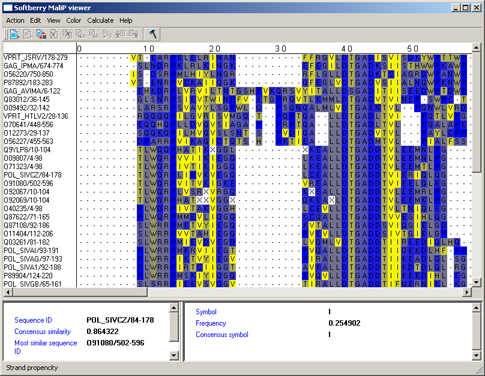 |
Figure 6. |
Turn propencity - preferring formation of the bending (fig. 7) [5].
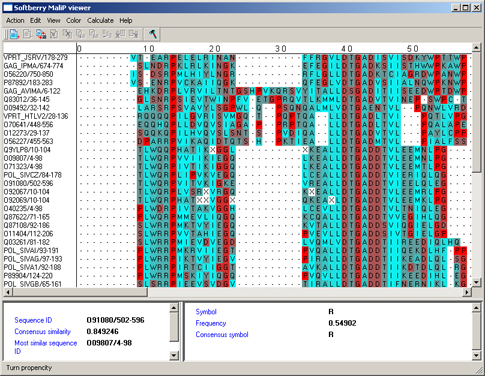 |
Figure 7. |
Buried index - in accordance to the frequency of occurrence inside a globule (fig. 3.3.8).
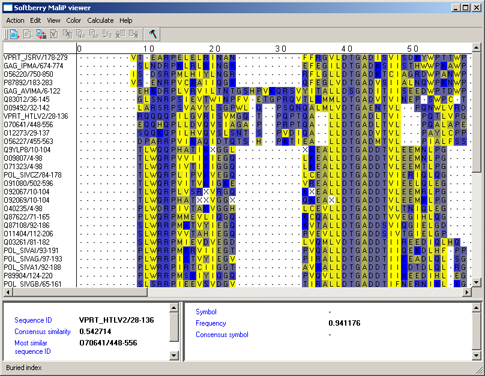 |
Figure 8. |
By conservation - it changes the color's intensity of used color scheme in dependence on conservatism [6,7]. The conservatism can be assigned with use of numbers 0 to 9 and a symbol "*". The number defines the number of characteristics that are to be similar for properties of aminoacids in the column of alignment. The "*" symbol means that the set of aminoacidic residues has the similar properties for all characteristics in consideration. For "*" the color cannot be changed. As for numbers, the lesser the number, the lighter the color. In figure 9 the result of serial using of "ClustalX colors" and "By conservation" schemes is shown.
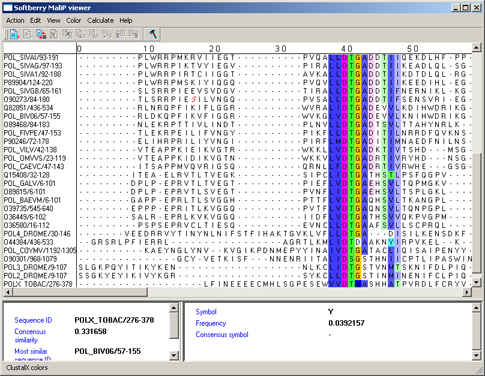 |
Figure 9. |
Above PID threshold only - the chosen color scheme (Zappo coloscheme, Taylor coloscheme
or By hydrophobicity) is to be applied only to those aminoacidic residues, percentage of which in the current
column exceeds the threshold value (fig. 10). This command opens the "Enter PID threshold" dialog (fig. 11),
where the user should define the threshold value and then press the "OK" or "Apply" button.
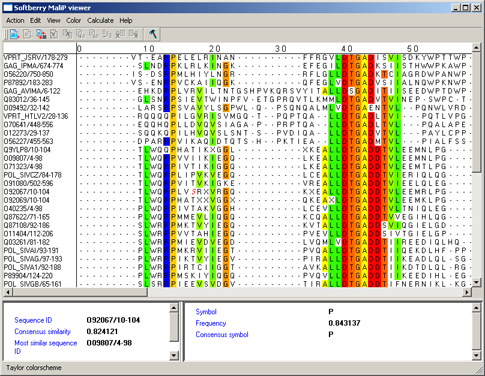 |
Figure 10. |
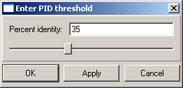 |
Figure 11. |
By PID - aminoacidic residues are colored in accordance to the rate of occurrence of a symbol in the column (see table 4, fig. 12). This scheme is used by default.
Table 4.
| Percent | Color |
| > 80 % | Mid blue |
| > 60 % | Light blue |
| > 40 % | Light grey |
| <= 40% | White |
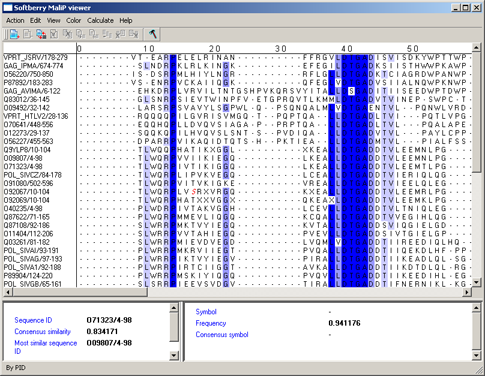 |
Figure 12. |
By BLOSUM62 score - aminoacidic residues are colored in accordance to their score in the column, which is determined accordingly to the BLOSUM62 matrix [1] (fig. 13). The symbol with maximal score is colored in blue. If the score of a symbol is positive when compared to the maximal one, it is colored in light blue, in all other cases the symbol is colored in white.
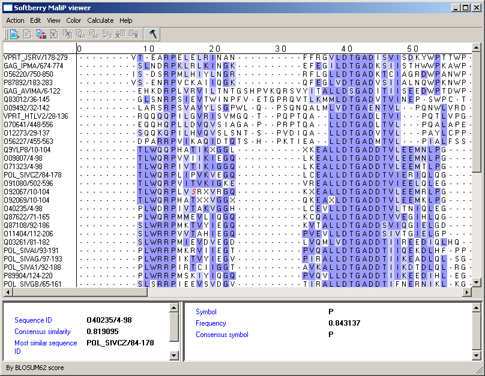 |
Figure 13. |