The Reads Mapping Viewer software is developed to visualize the results of reads mapping to a contig. It allows to operate large genomes as well as huge number of reads obtained using advanced full-genome sequencing technologies. The program allows to look through an alignment of reference sequence, assembled sequence, reads, spliced reads and sequence annotation at different scaling.
!Note! The program operates large amount of data, which may cause delay in displaying. Please be patient.
 |
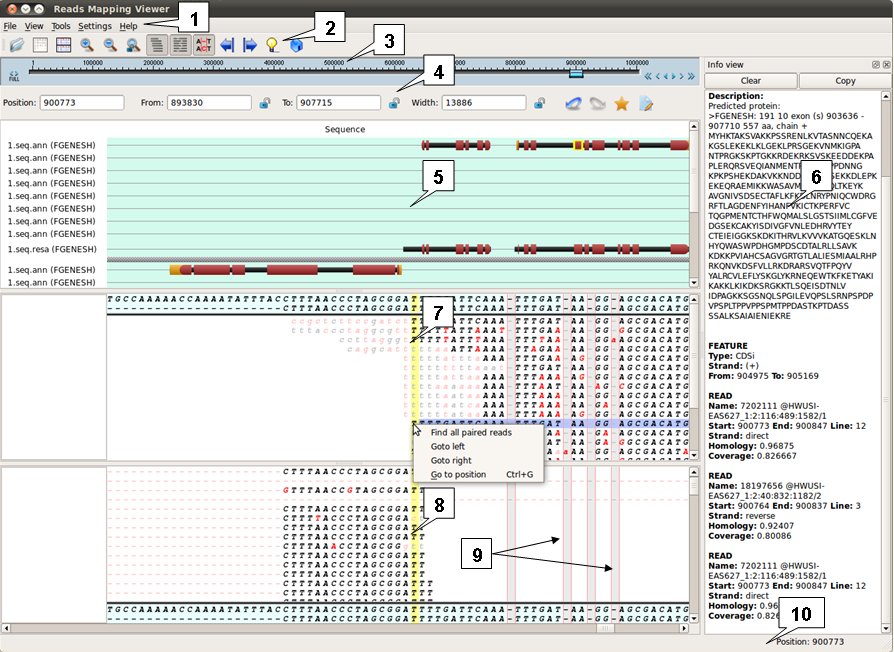
| 1. Main menu. 2. Toolbar. 3. Navigation scrollbar. 4. Navigation toolbar. 5. Structure view window. 6. Unspliced reads window. 7. Info view window . 8. Spliced reads window. 9. Inserts. 10. Status bar. |
Open reads — open file with reads.
Open annotation — open the annotation in xml-format(previously opened annotation will be automatically closed).
Add annotation — open the annotation file without closing the already opened annotations.
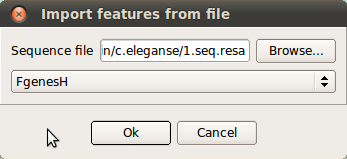
Import annotation — import the annotation file in FgenesH format without closing the already opened annotations.
Save image — save image from main window in .png format.
Close all — close all opened data.
Exit — quit the program.
Show inserts — display inserts in reads and corresponding gaps in reference sequences in the Unspliced reads window and Spliced reads window. If this option is disabled, inserts are not displayed and their presence is indicated by a vertical red line.
Save bookmarks to file — save current bookmarks into a file in plain text format, each position number is saved in a separate file row.
Load bookmarks from file — load bookmarks from a file.
Lock zoom — bind scaling and position in the Structure view window to Unspliced reads window and Spliced reads window. This option allows to control Unspliced reads window and Spliced reads window using Navigation scrollbar and Navigation toolbar. When Lock zoom is on, Scaling in Structure view window is disabled.
Zoom in, Zoom out — change scaling in the Structure view window.
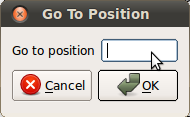
Go to position — jump to a specified position in the Structure view window, Unspliced reads window and Spliced reads window.
Unsplit structure view — remove all additional Structure view windows.
Split structure view — add an additional Structure view window. It may be convenient when comparing annotation rows that are located too far away from each other.
Show unspliced reads — display the Unspliced reads window, option is enabled by default.
Show spliced reads - display the Spliced reads window, option is enabled by default.
Info view — display the Info view window.
Find reads by name – search for reads by name.
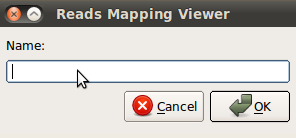
Find reads by index – search for reads by index.
Produce an annotation for the contig - generate an annotation in FgenesH format for a loaded contig.
Motif search — search for motif in a reference sequence. User can specify a motif itself as well as maximum number of allowed mismatches.
Set font – open the “Select Font” dialog.
Content... - show the Help window
About — show the program info window
Visit Website — link to http://www.softberry.com/berry.phtml
Buttons at the Main toolbar except "Previous border" and "Next border" duplicate the often used menu items. These buttons become active after the user had selected any feature in the Structure view window, and are intended for convenient navigation over reference sequence annotation. By pressing the “Previous border” or “Next border” button, the Position value is changed to that of the nearest left or, respectively, right border of a feature in a selected row. The current feature itself is highlighted yellow.
| File > Open | View > Unsplit structure view |
| View > Split structure view | |
| View > Zoom in | |
| View > Zoom out | |
| View > Lock zoom | |
| View > Show inserts | |
| View > Show unspliced reads | |
| View > Show spliced reads | |
| Go to previous border | |
| Go to next border | |
| Help > About | |
| Help > Visit Website |
Navigation scrollbar represents the location of visible area of the Structure view window in relation to reference sequence.
The ![]() button allows to rescale a sequence up to window size.
button allows to rescale a sequence up to window size.
| Shift the visible area to the left by the Structure view window width | |
| Shift the visible area to the left by 20% of the Structure view window width | |
| Shift the visible area to the left by 5 nucleotides | |
| Shift the visible area to the right by 5 nucleotides | |
| Shift the visible area to the right by 20% of the Structure view window width | |
| Shift the visible area to the right by the Structure view window width |
Position — when a number is entered, it is the same as “View > Go to position”, when the mouse pointer is moved in Structure view window, Unspliced reads window and Spliced reads window, the current position is displayed.
Fields From, To and Width control viewing in the Structure view window only. One of these parameters (From, To or Width) can be
locked by pressing the ![]() button. Once it is done, all other specified parameters need to be recalculated with regard to the locked parameter.
To apply changes, press Enter.
button. Once it is done, all other specified parameters need to be recalculated with regard to the locked parameter.
To apply changes, press Enter.
If the "Lock zoom" option is enabled, the scaling in Structure view window cannot be changed from the Navigation toolbar.
In the right part of Navigation toolbar there are buttons for managing bookmarks.
| Jump to previous bookmark. | |
| Jump to next bookmark. | |
| Set a bookmark. | |
| Open "Save bookmarks" dialog. Manage bookmarks: view the list of bookmarks, obsolete bookmarks can be deleted, you can jump to selected bookmark. |
Saving and loading bookmarks can be performed through the View menu items.
Manage bookmarks: view the list of bookmarks, obsolete bookmarks can be deleted by “Remove” button. To jump to bookmark select it and press "OK" button. To the list at the top part of the window, the search results (search by name or index, or search for paired reads) are being committed.
 |
In this window, top upper row shows the reference sequence, the next row represents an assembled sequence, all following rows represent reads.
Positions, in which an assembled sequence and/or read(s) differ from the reference sequence, are marked red. Non-significant reads' positions are pale and in lowercase. The information on this read is displayed when the mouse pointer hovers over a read.
Same rules as above are used to display reads in Spliced reads window. The consensus is shown in the last row, and the contig is shown in the row before that.
Mouse click inside Unspliced reads window or Spliced reads window causes yellow highlighting of a column that corresponds to current position of the mouse pointer. The indicated position in the Structure view window is being marked by a thin vertical line. If necessary, information is automatically scrolled to move the position into visible area.
Scrollbar located below the Unspliced reads window and Spliced reads window allows to scroll the information in these windows independently, without correspondence with the Structure view window.


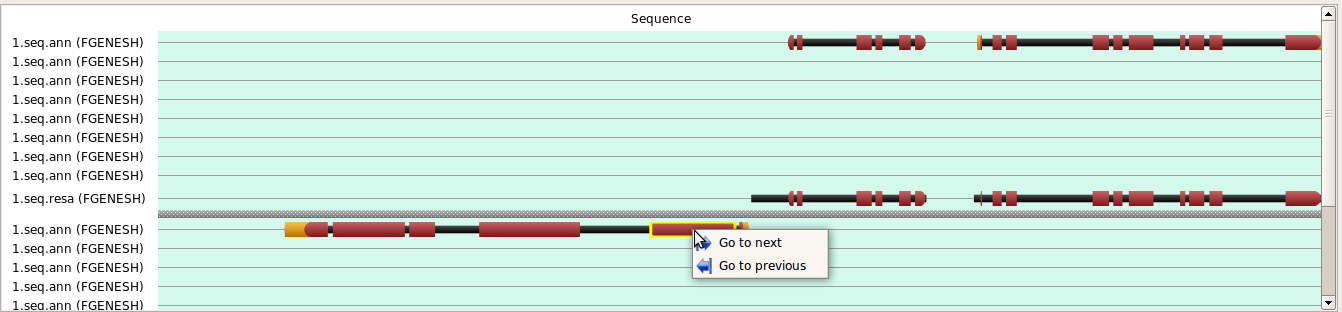
The window displays loaded annotation. Feature can be selected by mouse (selected feature is highlighted yellow). «Previous border» and «Next border» is used to navigate over features. If needed, the window can be split using «View > Split structure view». Current position, if it is inside a visible area, is shown by a vertical line.
The window displays information on a feature selected in the Structure view window or on selected read.
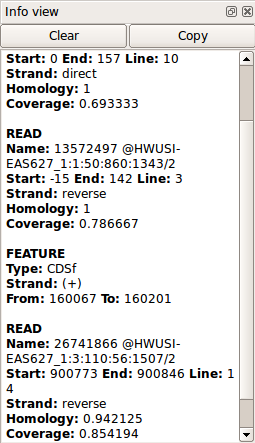
Displays the current cursor position.