Services Test Online
AbSplit - Separating archea and bacterial genomes
ABSPLIT is a program that recognizes the belonging of bacterial DNA to one of two realms: Bacteria or Archeabacteria. The algorithm of recognition is based on calculation of linear discriminant function on 88 criteria. 84 criteria correspond to frequencies of 1-2-3 nucleotides, 2 criteria correspond to maximal lengths of AT and GC tracts, and the last 2 criteria are the coefficients of linear correlation of codons' frequencies in ORF of maximal length in a test sequence with codons' frequencies in genomes, which belong to Archeabacteria and Bacteria correspondingly. If a value of linear discriminant function is more than 0, then a sequence belongs to Archeabacteria's realm, otherwise - to Bacteria's realm.
As the input data, DNA sequences in FASTA format are used, and for each sequence a score is calculated. The total statistics on set of sequences is placed in the beginning of output file (numbers and parts of predicted sequences, related to different realms). Further, the histogram for distribution of linear discriminant function's values in a set of sequences is shown. And after this, the classified sequences, whether they are bacterial or archeabacterial, are shown.
Analysis of the test data (53399 sequences of 97 bacterial/archeabacterial genomes) revealed the preciseness of classification (the rate of correctly identified sequences) equal to 0.886.
OUTPUT EXAMPLELDF discrimination threshold=0.000000 Prediction results: Number of sequences=129 Arch(num/fract)=64/0.496124; mean_score=1173110.225735 Bact(num/fract)=65/0.503876; mean_score=-679245.160401 Histogram: 1 -1653112.270017 -1492294.115256 0.007752 2 -1492294.115256 -1331475.960496 0.015504 3 -1331475.960496 -1170657.805735 0.015504 4 -1170657.805735 -1009839.650974 0.038760 5 -1009839.650974 -849021.496214 0.069767 6 -849021.496214 -688203.341453 0.085271 7 -688203.341453 -527385.186693 0.093023 8 -527385.186693 -366567.031932 0.108527 9 -366567.031932 -205748.877172 0.023256 10 -205748.877172 -44930.722411 0.038760 11 -44930.722411 115887.432349 0.031008 12 115887.432349 276705.587110 0.054264 13 276705.587110 437523.741870 0.015504 14 437523.741870 598341.896631 0.023256 15 598341.896631 759160.051392 0.062016 16 759160.051392 919978.206152 0.023256 17 919978.206152 1080796.360913 0.015504 18 1080796.360913 1241614.515673 0.038760 19 1241614.515673 1402432.670434 0.046512 20 1402432.670434 1563266.457703 0.038760
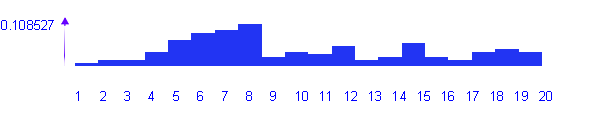
Predicted archaeal sequences: >AB001339|seq56|1 ttagtcagggggccccgccgatgaaaccggggacagctactaaacccattgccagtggtgg tggtagctctggccctagtctgggctccggccaacccagagcagaacggcccggtggcggc aatgcaggggcaaatgttggtcccattgcggccaatcccgttgctagtagtgctcccccta aaccgaaaccaactcccagttcccccgctaagccagaccccttaaagtgcgttagccaatg taaacccagttatccctccatcctccagggggaagaaggtagtgctacagtattaatttca gtaaatgatagtggtggtgtgaccagcgtaaccatcaccaatgcccacggcaacagcgagg tcaaccgccaggccctattggcagccagaaaaatgcagtttacggcccccgccagtggtca atccaaatcagtccctgtggtgattcacttcaccgttgctggttcagactttgatcgtcag gcgagggagcgtcagcaacagcaggaagagttgcgtcaggccgcccgcagagcagaagagg aaaaggcaaatcaagcccgtcagagacagttggaagaggagcgtcaagcccgccaacggca attagagaaagaacgggaag >AB001339|seq128|1 aggcttccaagcaagcttcaattaaggatttttccagaaagggatcccccacctgcaccgc tgggcgatcgtccatggactgatccgttaactcagcactggcaaaactggctccccccatg ccatcccgtcccgtggtggaaccgacatataaaactggattgcctatcccagaagccccag ctttgacaatttcttccgtttccatcaaacccaaggccatggcgttgacgaggggattacc ggagtaagccggatcaaagtagatttccccgcccacagtgggcacaccaacacaattaccg taatgactgatcccatccactaccccggtgaaaatacgtcgattcctagcatcgtccaaat taccgaaccgtagggaatttaaaatggcgatcggcctcgctcccatggtgaaaatatcccg cagaatcccccctactccggtggcggctccctggaatggctccactgcggaaggatggtta tgggattcgattttaaacgccaatctcaggccatcccccaaatctacgaccccggcatttt ccccaggccccactaaaatgcgttctccttcggtgggaaagttactcagtaggggacggga atttttataacaacaatgtt >AB001339|seq184|1 attttcccgaagaaactacctccgatgcttggctgaccccagcagatgccggccaggatgg tgatgcccaggaaccggcggaagatgggggagaagaaggagtagtgtcggaagaactggcc ctgcctgaggacttacctcctatggatgccatggtggcggcagtggaagaaatgactccgg tggtggtgcccgaaactgtaccagaaacagaaaccccagccttagaggatttggtcgccca aaagaccgccctggaaaaggacattgccgctctgcaacgggaaaaagcccagtggtatggc cagcagttccagcaattacagcgggaaatggcccggttagtggaggaaggcaccagggaat tagggcaaagaaaagcagctctggaaaaggaaattgagaagttagagcgccgtcaggaacg gattcaacaggaaatgcgtaccacttttgccggggcttcccaggagttggccatccgcgtg cagggctttaaggattatttggtggggagtttgcaggatttggtttccgccgccgaccagt tggaattaggggtgggggacagttgggagtcttcctctacccatggggatgcgattattga aaatgccgacccaactccgg >AB001339|seq336|1 tctgccagctttgccattaatttccgcctcgatcccaccgaggtcgttaccattcgccgca cccaaggcacgttacaaaatattgtcgccaagattattgctccccaaacccaggaatcttt taaaattgccgccgcgcgacgcacagtggaagaagccatcaccaaacggagcgagttgaag gaagactttgataacgcccttaattcccgcctggagaaatacggcatcattgttctggaca ccagtgtggtggatttagccttctcccccgaatttgccaaggcggtggaggaaaaacaaat tgctgagcagagagcccagcgggcagtgtatgtggcccaggaagcggaacaacaggcccag gcggacatcaaccgagccaaggggaaggcagaagcccaacggttactggcggaaactttaa aagctcaggggggggaattagtcctacaaaaagaggcgatcgaagcttggcgggaaggggg ggctcccatgcccaaggttttggtgatggggggagaaggcaaggggtctgcggttcccttt atgtttaacctaactgacctggctaactagcggcagcggggaagttataggtcccagggct cctgcctgacctttaggtcc … Predicted bacterial sequences: >AB001339|seq8|1 ctgttacgtgttttgttgcaaacggaactttttgcagtagttagctccgttgttgccgata ccagtcaatggtatttttcaatccttcccgcaagctcacctgggcttcaaacccaaattct gctttagctttggtggtgtctaaacagcgacggggctggccgttgggttgatcggtttccc aaataatgtccccctcaaactccatcagttcacagattaattccgttaagtctttgatgga aatttcaaaattggtgcctaggttaaccggatcggctttgtcgtaggcttgggttcccatc acaatgccccgggccgcatcagtggagtaaagaaattccctggtgggactgccgtcgcccc aaacgggtaattgtttttgtccagctttttgcgcttcgtaaaccttatggatcaaggcagg aatcacgtgggaactgcggggatcgaagttatcttctgggccgtaaagatttactggcaag aggtaaatgccattaaagccatactgcaagcggtaggattccagttgcaccaacaatgctt tcttggccacgccgtagggagcgttggtttcttcaggataaccgttccataagtcttcttc cttaaagggtacaggggtaa >AB001339|seq24|1 cctttttttatttatcttgcccgctcccaaattaaataatcaaacctaacgggtcaactcc aaagacaacccaaggccattccaggctaattgattgaatcccgaattttattaactgtttg ttccatttgtgccatgtttgcccctcgaccttggattgtggtccgtctccggtctttaccc ctatcgtttcgcctcgatcgccatgtccccttggtaatgggattacttactgctctagcat tattactatttattctcaatattagttggggggaatatcctgtccctcccttggcgatgct ccaggccatctttgggctatctaccgatgctgaccatgaatttgtggtgcgtactctgcga ttaccccggtccttggtggcattgttggtgggtatgggtttggcgatcgccggagggattt tgcaaggcattacccgcaatcctttggcagcccctgaaattattggtgtcaatgcgggggc tagtttggtggcggttaccttcatcgttttgctaccgggtatttctccttccttgctgcca gtggccgctttttgcggtggtttaacagcggcgatcgccatttatgtgctggcttggaatc agggcagtgcccccgtccgg >AB001339|seq32|1 atgatgttgattactcctccagtggcaccatccccgtaaatggccgttggcccctggatca cttcaatccgttcaatggcactgggagcaatggtttgcaaatctcggaaggcattacggtt ggtggtttggggcacaccgtcaatcaaaaccaaaacgttacgtcctcgcaaagcctggcca aattgactggcactcccggtgctgggggctaagcctggcactagttgacccaaaatatccg ccaaggaagagtaaacctgggtttgttgctcaatttctgcccgttcaattaccgttaccga ccggggaatgttagcgatttcctcctctgtacgggtggcggaaaccacaatttgtagggcc tcactttcctctatctcggcggttgtcccggcaacccctggtcgaatcagcaattgtaacc cttgcgagttaggctttacttcggcttccggtggcccatttacccccgtgatagctaagcg cacttggttatcggtcatttgggtaacactgacaaacgcaatgtccgcagtggggctcact tcttcaaacccctggcccccaggtaaggccatcaaagtattgggaagatcaataattaagg cattgcccaccgtttgtagg >AB001339|seq64|1 ccgtccccgtcttaccggtaaagtatttgagaattagttgcagttaaggttgttcctcctg tgttatcagatgccatggccggctgtctcaactaagaatttcaagctttggtgcaaggagt gattatgaatcaagtacagtggtcggttttgttgatgggtatagtttcgctactatgtgct cccagggcgtgggccgaaactaatccgaaccaattgaacaggacgaatattttagaatctg gtaacttagaacgcaccaaagccggtgatttgctcccagttgcaaccactgttgatgagtg gataacccaaattgcccaagcttcgatcatcgaaatcaaggaagcccggatcaatttgacc gaagctggactggaactgaccctggctaccacgggccgcttatcaacaccaaccacttccg tagtgggcaatgcactaattgtagatattcccaatgccatcctagccttgccggatagtga cggactgcaacaggaaaaccccaccgaagaaattgccctagtgagcgttacagcattacct gataatattgttcgcattgccattaccggggtcaatgtgccgccgacggttgaagttaatg ccacagaccaatccctggta …