Services Test Online
FGENESH - Program for predicting multiple genes in genomic DNA sequences
FGENESH is the fastest (50-100 times faster than GenScan) and most accurate gene finder available - see the figure and the table below. In recent rice genome sequencing projects, it was cited "the most successful (gene finding) program (Yu et al. (2002) Science 296:79) and was used to produce 87% of all high-evidence predicted genes (Goff et al. (2002) Science 296:79).
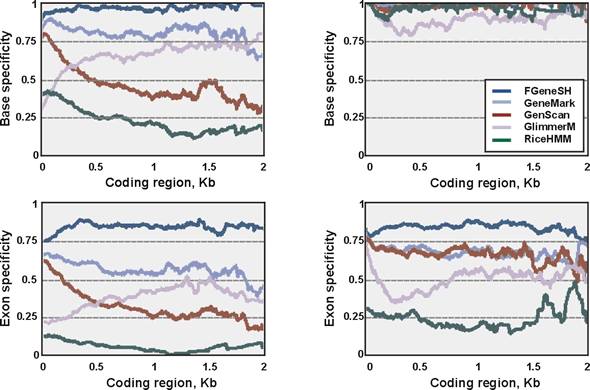
(above) Performance of different gene finding programs on rice genes. Reprinted from Yu et al. (2002) Science 296:79-92. FGENESH is by far the most accurate of five programs tested.
(Below) Performance of three popular gene prediction programs on 42 semiartificial genomic sequences containing 178 known human gene sequences (900 exons). Sensitivity is percentage of exons that are predicted correctly. Selectivity is percentage of predicted exons that are correct. Reproduced with changes from Yada et al., 2002 Cold Spring Harbor Genome Sequencing and Biology Meeting, May 7-11, 2002. FGENESH is by far the most accurate of three programs.
| Program |
Sensitivity
|
Specificity
|
Missed Exons, %
|
Wrong Exons, %
|
| FGENESH |
77.1
|
65.7
|
9.6
|
23.2
|
| GenScan |
66.5
|
44.9
|
12.0
|
40.9
|
| HMMGene |
69.6
|
36.6
|
15.5
|
55.5
|
Web version of FGENESH can be used with parameters for the following genomes: human, mouse, Drosophila, nematode, dicot plants, monocot plants, yeast (S.pombe) and Neurospora.
Check appropriate genome/organism and and FGENESH program. Paste your sequence to the window or load your file with sequence in FASTA format and click Perform Search button.
References: Salamov A., Solovyev V. (2000)
Ab initio gene finding in Drosophila genomic DNA.
Genome Res., 10,516-522
Fgenesh output:
G - predicted gene number, starting from start of sequence;
Str - DNA strand (+ for direct or - for complementary);
Feature - type of coding sequence: CDSf - First (Starting with Start codon), CDSi - internal (internal exon), CDSl - last coding segment, ending with stop codon);
TSS - Position of transcription start (TATA-box position and score);
Start and End - Position of the Feature;
Weight - Log likelihood*10 score for the feature;
ORF - start/end positions where the first complete codon starts and the last codon ends.
FGENESH-2.1 Prediction of potential genes in genomic DNA Time: Thu Jan 28 19:25:51 1999. Seq name: HUMHBB 73308 bp DNA PRI 20-JAN-1994 length of sequence 73308bp G+C content: 39 Isochore: 1 number of predicted genes 7 in +chain 7 in -chain 0 number of predicted exons 18 in +chain 18 in -chain 0 Gn S Type Start End Score ORF Len -- - ---- ----- --- ----- --- --- 1 + TSS 19447 -7.15 1 + CDSf 19541 - 19632 16.12 19541 - 19630 90 1 + CDSi 19755 - 19977 14.12 19756 - 19977 222 1 + CDSl 20833 - 20961 2.99 20833 - 20961 129 1 + PolA 21055 1.05 2 + TSS 34437 -7.15 2 + CDSf 34531 - 34622 15.25 34531 - 34620 90 2 + CDSi 34745 - 34967 20.74 34746 - 34967 222 2 + CDSl 35854 - 35982 5.59 35854 - 35982 129 2 + PolA 36043 1.05 3 + TSS 39373 -7.15 3 + CDSf 39467 - 39558 15.25 39467 - 39556 90 3 + CDSi 39681 - 39903 20.74 39682 - 39903 222 3 + CDSl 40770 - 40898 5.74 40770 - 40898 129 3 + PolA 40959 1.05 4 + TSS 44415 -8.75 4 + CDSf 45995 - 46151 16.01 45995 - 46150 156 4 + CDSl 46997 - 47100 2.71 46999 - 47100 102 4 + PolA 47243 1.05 5 + TSS 54703 -4.45 5 + CDSf 54790 - 54881 13.41 54790 - 54879 90 5 + CDSi 55010 - 55232 14.20 55011 - 55232 222 5 + CDSl 56131 - 56259 3.87 56131 - 56259 129 5 + PolA 56365 1.05 6 + TSS 62100 -6.65 6 + CDSf 62187 - 62278 13.59 62187 - 62276 90 6 + CDSi 62409 - 62631 19.50 62410 - 62631 222 6 + CDSl 63482 - 63610 10.23 63482 - 63610 129 6 + PolA 63718 1.05 7 + TSS 68088 -9.45 7 + CDSo 68183 - 68428 14.87 68183 - 68428 246 7 + PolA 68509 1.05 Predicted protein(s): >ID 1 3 exon (s) 19541 - 20961 147 aa, chain + MVHFTAEEKAAVTSLWSKMNVEEAGGEALGRLLVVYPWTQRFFDSFGNLSSPSAILGNPK VKAHGKKVLTSFGDAIKNMDNLKPAFAKLSELHCDKLHVDPENFKLLGNVMVIILATHFG KEFTPEVQAAWQKLVSAVAIALAHKYH >ID 2 3 exon (s) 34531 - 35982 147 aa, chain + MGHFTEEDKATITSLWGKVNVEDAGGETLGRLLVVYPWTQRFFDSFGNLSSASAIMGNPK VKAHGKKVLTSLGDAIKHLDDLKGTFAQLSELHCDKLHVDPENFKLLGNVLVTVLAIHFG KEFTPEVQASWQKMVTGVASALSSRYH >ID 3 3 exon (s) 39467 - 40898 147 aa, chain + MGHFTEEDKATITSLWGKVNVEDAGGETLGRLLVVYPWTQRFFDSFGNLSSASAIMGNPK VKAHGKKVLTSLGDAIKHLDDLKGTFAQLSELHCDKLHVDPENFKLLGNVLVTVLAIHFG KEFTPEVQASWQKMVTAVASALSSRYH >ID 4 2 exon (s) 45995 - 47100 86 aa, chain + MGNPKVKAHGKKVLISFGKAVMLTDDLKGTFATLSDLHCNKLHVDPENFLVSTLRQRDID CFGNPLQRGFYPTDTGFLAVTNKCCG >ID 5 3 exon (s) 54790 - 56259 147 aa, chain + MVHLTPEEKTAVNALWGKVNVDAVGGEALGRLLVVYPWTQRFFESFGDLSSPDAVMGNPK VKAHGKKVLGAFSDGLAHLDNLKGTFSQLSELHCDKLHVDPENFRLLGNVLVCVLARNFG KEFTPQMQAAYQKVVAGVANALAHKYH >ID 6 3 exon (s) 62187 - 63610 147 aa, chain + MVHLTPEEKSAVTALWGKVNVDEVGGEALGRLLVVYPWTQRFFESFGDLSTPDAVMGNPK VKAHGKKVLGAFSDGLAHLDNLKGTFATLSELHCDKLHVDPENFRLLGNVLVCVLAHHFG KEFTPPVQAAYQKVVAGVANALAHKYH >ID 7 1 exon (s) 68183 - 68428 81 aa, chain + MEQSWAENDFDELREEGFRRSNYSKLKEEVRTNGKEVKNFEKKLDEWITRITNAQKSLKD LMELKTKAGELRDKYTSLSNR