Services Test Online
pairDistance - Programs for collect, analyze and visualization pair-ends reads distance.
CONTENTS
Description
Synopsis
Commands And Options
License And Citation
Downloads
Description
Here we describe the programs for collecting, analyzing and visualizing distances within paired-end reads. Pair_distance program attempts to align reads on contigs and collects statistical data on distance between reads. Then, a user can load that data into visualization program SView than select a region on a curve and get parameters of distribution.
SYNOPSIS
To collect information about distance in paired-end reads.
./bin/pair_distance chr13.fa reads.fa -o:pd.cfg
To display distribution chart of distances (Fig. 1).
./SView/Sview -type PE
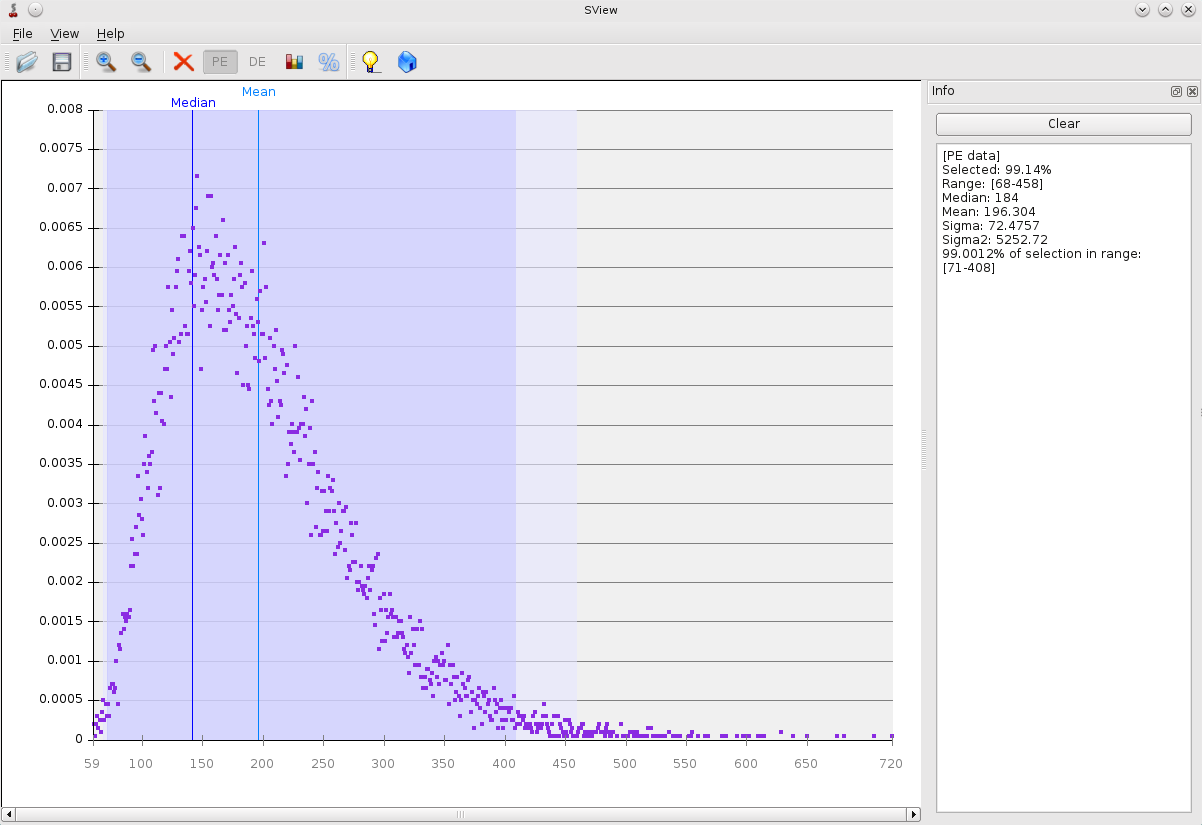
Fig. 1 Result of the visualization.
Commands And Options
./pair_distance contig.fna reads.fna -o:config [options],
where:
contig.fna - fasta file with one or more genomic contigs.
reads.fna - fasta file with paired-end reads, sequences of paired reads should be in order even/odd.
pair_distance options:
-PE_only - selects data for "PE" reads only
-MP_only - selects data for "MP" reads only
-DE_only - selects data for "distance ends" reads only with very long distances (>1K bp) between ends.
A name of an output file is the same as a name of input with added “.distance” extension.
./SView -type PE|MP|DE
Sview
is a QT-based GUI program for analysis and visualization of distance data on paired-end reads. User must specify type of paired reads before loading a data file.
-type PE|MP|DE - specifies reads type.
-in "data.bin" - loads file with data in binary format. Usr can also specify a datafile in menu "File"
-out "result.txt" - specifies name of an output file. If name isn’t specified, an output will be printed in stdout.
User can navigate a distribution curve with a mouse wheel and use Ctrl-mouse wheel to zoom in and out. Statistic data is printed in an info window if a representative part of curve is selected by clicking and dragging a mouse pointer. For "PE" and "DE" reads, user must select a single area. In case of “MP” reads, user can switch between types of area by pressing "PE" and "DE" buttons in a toolbar. User can save an output by pressing a button on a toolbar or by using "File>Save" menu command.
License And Citation
pairDistance is free for academic usage. For commercial licensing, please inquire to softberry@softberry.com.
Downloads
pairDistance for academic usage can be downloaded here