Services Test Online
Proteomics-MSSmoothing - Softberry Mass Spectra (SMS) processing tools. Data smoothing.
Data smoothing procedure is intended for data noise elimination. During the smoothing, the values of intensity for each mi point are being averaged by several neighboring points. The number of such points is determined by the 'SmoothWindowSize' parameter (default value is 3). The smoothing procedure can be repeated for several times; the number of iterations is determined by the 'SmoothReps' parameter (default value is 3). Example of data smoothing is shown in the figure 1.
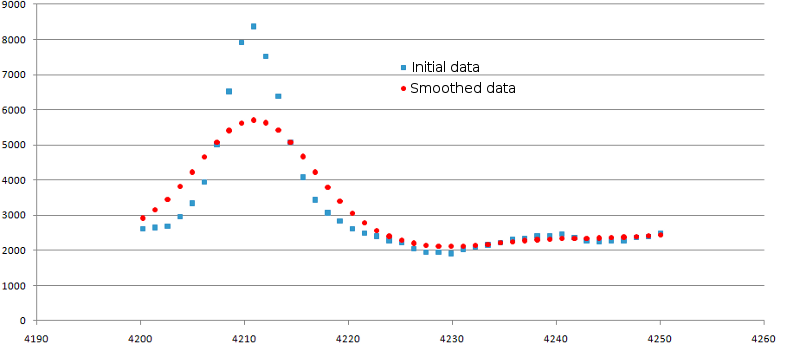
Figure 1. Result of data smoothing. Original data are shown as blue squares, smoothed ones - as red circles.
The SmoothWindowSize was set to 3 and SmoothReps was set to 3.
Input: m/z - Intensity data
Output: Smoothed m/z - Intensity data in the same format as input data.
Parameter(s):
SmoothWindowSize - This parameter determine window size for smoothing operation. The default value is 3.
SmoothReps - This parameter specify the number of smoothing operation repeats. The default value is 3.
File format type - This parameter specify file format. SSV-space separated values, CSV - comma separated values, TSV - tab separated values.
Data format.
Mass spectra data represent the sets of following pairs of values: mass to charge relation
(m/z, further, for more convenience, it will be referred to as m, mass) and corresponding signal intensity (I).
On a spectrum plot, the mass corresponds to X coordinate, and signal intensity- to Y one.
A typical spectrum consists of several thousand of such value pairs (points). Data are represented as
text files, where for each pair (mi,Ii) of mass-intensity values the string is assigned, and data in
this string are separated by special separator symbol. The SMS package allows several separators types:
space (SSV, space separated values, file format), comma (CSV, comma separated values, file format) and
tabulation (TSV, tab-separated values, file format). In files with data, the string with comments are
allowed; during the file reading these strings are to be skipped. The commentary strings should begin
with "#" symbol at the first position. In the figure 2 the example of file with data in CSV format is shown.
#M/Z,Intensity -7.8602611e-005,4.1126194 2.1773576e-007,4.0764203 9.6021472e-005,4.0040221 0.00036601382,4.1186526 0.00081019477,4.0040221 0.0014285643,3.9617898 .... 19742.941,4.077895 19745.564,4.0772248 19748.187,4.0772248
Figure 2. Example file with mass spectra data in CSV format.