Services Test Online
Transcripts Assembler (TRA) - Assembly of transcripts from RNASeq reads (without genome sequence information)
Description
Quick-start
Build
Commands and options
Configuration files
License and citation
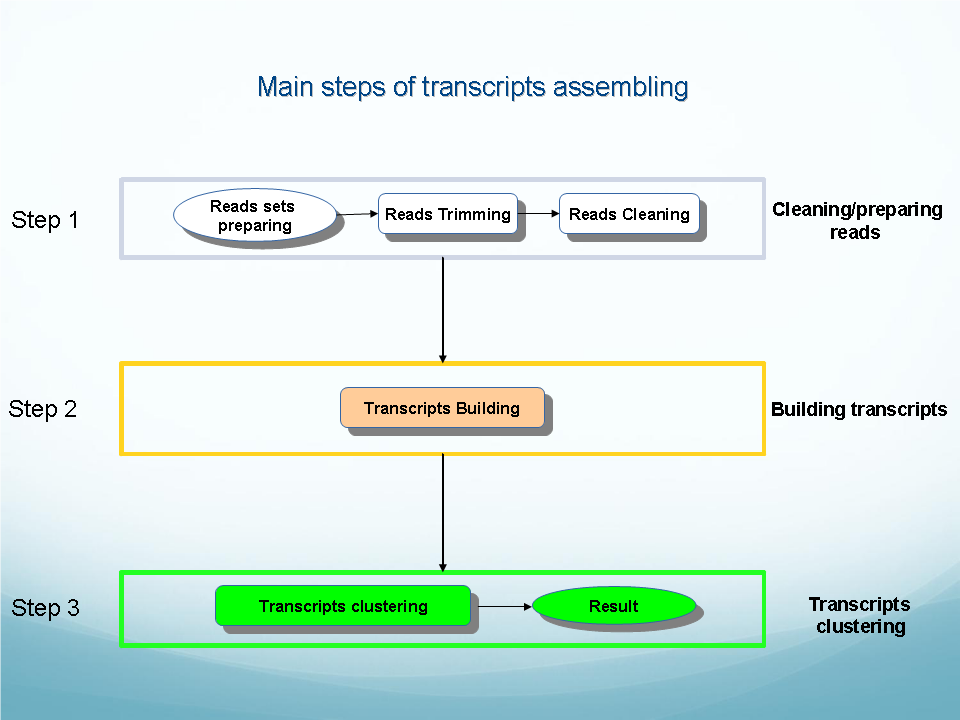
|
| 1. Cleaning and preparing reads. 3. Building transcripts. 4. Clustering transcripts. |
Description
TranscriptAssembler is a software package for assembling transcripts from sets of small reads.
Quick-start
Run the assembly:
ReadsAssembler4/assembler.pl -cfg:ReadsAssembler4/cfg/assembling.def.cfg -cfg:./user.cfg -work_dir:./wrk -j:8
The result will be saved in ./wrk/res folder
Source data are specified in the configuration file user.cfg (you can edit file user.cfg in a sample folder):
reads.pe.src {
…
src.file = "reads.fa"
src.format = "ifasta-pair"
aver_pe=200
stdev_pe=30
}
Build
In order to compile the software package, the following script shall be executed in ./src folder. Command line: make_all.pl --release.
Commands And Options
Example of command line:
ReadsAssembler4/assembler.pl -cfg:ReadsAssembler4/cfg/assembling.def.cfg -cfg:./user.cfg -work_dir:./wrk -j:8
Specify configuration files
cfg:fname - Main settings are specified in configuration files. Default settings are stored in cfg/assembling.def.cfg, this file must be entered in a command line before users' own configuration file. User can also create and use his own configuration file(s) (user.cfg in an example), which would contain paths to input files and user-defined (see section CONFIGURATION FILES). User-specified configuration file must always follow default file in a command line.
Working folder.
-work_dir:XX - Specifies working directory. Results are saved in subfolder XX/res.
Multiprocessing.
-j:XX - Number of processors that the software package is permitted to use
Configuration Files
Source data
Input data are specified in user configuration file, which can contain one or several sections. Each read set is described in its own separate section of that file.
Lines starting with symbol # contain comments.
Below is an example of a section specifying input reads, with:
reads.pe.src1{
--trimming
--cleaning
--assembling
--scaffolding
src.file = "reads.fa"
src.format = "ifasta-pair" {
def_fasta_qua = 25
phread=64
}
trimming {
--adapters_trim
--cut_polyN
--cut_qual
adpt1_seq = ["AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC"]
adpt2_seq = ["AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGCCGTATCATT"]
}.
# --clean-by-self:off
# --clean-other:off
# --clean-by-other:off
aver_pe=200
stdev_pe=30
}
Name of each section, which specifies characteristics of an individual read set, consists of three identifiers separated by a period (.) First identifier for this type of section is always "reads".
Next identifier defines type of pairwise reads:
.se - "SE" reads type – single reads
.pe - "PE" reads type – "PE" pair-end reads
.mp - "MP" reads type – "MP" pair-end reads
Third identifier is a unique name of a given set. Any combination of letters and numbers can be used. So, a section with name reads.pe.src1 defines parameters for "PE" pair-end reads.
Keys
Each section specifies keys that determine the parameters of operation with reads. The keys assume values "on" and "off". If a value is missing it is assumed to be "on". Therefore, record –cleaning:on is equivalent to а –cleaning.
Trimming
Trimming of a set is turned on by a key –trimming.
Trimming, i.e. preparing the reads for assembly, can include trimming adapters, polyN and tail sequences of low quality. Trimming parameters are specified in subsection trimming {} separately for each read set.
trimming {
--adapters_trim
--cut_polyN
--cut_qual
adpt1_seq = ["AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC"]
adpt2_seq = ["AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGCCGTATCATT"]
}
--adapters_trim Turns on trimming of adapters.
--cut_polyN Cut polyN mode. Search and remove polyN tails.
--cut_qual Cut by quality mode. Search and remove tails with bad quality.
-adpt1_seq:["..." "..."] Read1 adapter sequence, may be set more than once
-adpt2_seq:["..." "..."] Read2 adapter sequence, may be set more than once
Cleaning
Cleaning of a specified read set is turned on/off by key –cleaning.
ReadsAssembler performs preliminary cleaning of reads to get rid of random errors. By default, all read sets with cleaning turned on are used to clean each other. This behavior is regulated by three keys, which are turned on by default but can be toggled off for each read set if desired.
--clean-by-self:on – clean-by-self:on – Cleans read set using reads from the same set. To turn on: --clean-by-self:off.
--clean-by-other:on - Clean given read set using reads from other sets. To turn off, enter --clean-by-other:off
--clean-other:on - Clean other read sets using reads from this set. To turn off, enter --clean-other:off
Keep in mind that MP reads aren’t used for cleaning PE reads, but PE reads are by default used to clean MP reads. This setting is recorded in a default configuration file and cannot be changed by user.
As a result of cleaning, reads are sorted into two categories:
1. Reliably cleaned ("clean subset") – percentage of errors is reliably low.
2. Reads not reliably cleaned ("dirty subset") – reads that cannot be verified and/or cleaned along their entire lengths
(due to low coverage multiple and/or high rate of errors). Accordingly, the percentage of errors in this subset is very high.
--single-out:on – Save all results in a single file, i.e. not separate them into files containing cleaned and "dirty" subsets.
If this key is turned off (default setting) only reliably cleaned reads are saved - and sorted into two files:
identifier .pe.pair – Cleaned reads that retained a pair (both reads of a pair were reliably cleaned).
identifier .se.single – Cleaned reads that lost their pair (only one read was cleaned reliably).
If this key is turned on (--single-out:on), all reads - reliably cleaned and “dirty” - are saved in the same file.
Source data description
- src.format = "fasta-pair" – Format of a file containing reads. Allowed formats:
- fasta-pair –FASTA file where a second read for each pair (even) immediately follow a first (odd).
- fasta – for sections of se type, FASTA file with SE reads. For sections of pe or mp types, two files shall be specified, where first reads for each pair are recorded in a first file, and second reads – in a second file in the same order.
- fastq-pair – FASTQ file with pairwise reads directly following each other (even, odd).
- fastq – for sections of se type, FASTQ file with SE reads. For sections of pe or mp types, two files shall be specified, where first reads for each pair are recorded in a first file, and second reads – in a second file in the same order.
src.file = "reads.fa" – defines path to file(s) with reads (either absolute or relative paths). For pairwise reads separated in two files, two paths shall be entered inside square parentheses: src.file = ["reads1.fa" "reads2.fa"].
aver_pe=200 – Average distance between reads.
stdev_pe=30 – Standard deviation of that distance.
Transcript assembly
Use of specific base of reads in transcript assembly is turned on by key ––transcripts.
Parameters for transcript assembly are specified in a separate section transcripts {}, below is a sample of such section with explanation of keys:
transcripts {
min_seq_len = 500
result = "transcripts.fna"
}
min_seq_len = 500 – minimum length of assembled transcripts.
result = "transcripts .fna" – Name of an output file.
Hardware
Section Hardware sets size of memory in GB (whole numbers only) that can be used by ReadsAssembler:
hardware {
memory.opt = 4
memory.max = 8
}
Parameter memory. max sets the size of memory, and parameter memory.opt – optimum size of memory available for programs of ReadsAssembler..
Description of Program Output
The program output is saved in the following fasta files:
transcripts.fa: contains all assembled transcripts;
transcripts.fa.trash: contains discarded transcripts;
transcripts.fa.clust: contains transcripts grouped into clustered;
transcripts.fa.best: contains best transcript for each cluster.
Sequence identifiers in these files consist of three numbers. The first number is an ordinal number of a transcript, the second – its cluster number, and the third is the number of transcript in that cluster. For example,
> 5 4 2 -
or
> 1:1228 1
For each fasta file of an output, a corresponding file with some statistics is created:
transcripts.fa.cov
transcripts.fa.trash.cov
transcripts.fa.clust.cov
transcripts.fa.best.cov
Files with names starting with pe* contain sequences of cleaned reads.
LICENSE AND CITATION
Transcripts Assembler is free for academic use. For commercial licenses, please contact Softberry by e-mail to softberry@softberry.com.