Services Test Online
Transcripts Assembler (TRAG) - Assembly of transcripts from RNASeq reads (with genome sequence information)
SYNOPSIS
DESCRIPTION
COMMANDS AND OPTIONS
CONFIGURATION FILES
LICENSE AND CITATION
DOWNLOADS
|
SYNOPSIS
Assemble reads into transcripts.
perl reads_assembler/assembler.pl -work_dir:./wrk -j:8 -cfg:reads_assembler/cfg/assembling.def.cfg -cfg:./reads.sim.cfg
DESCRIPTION
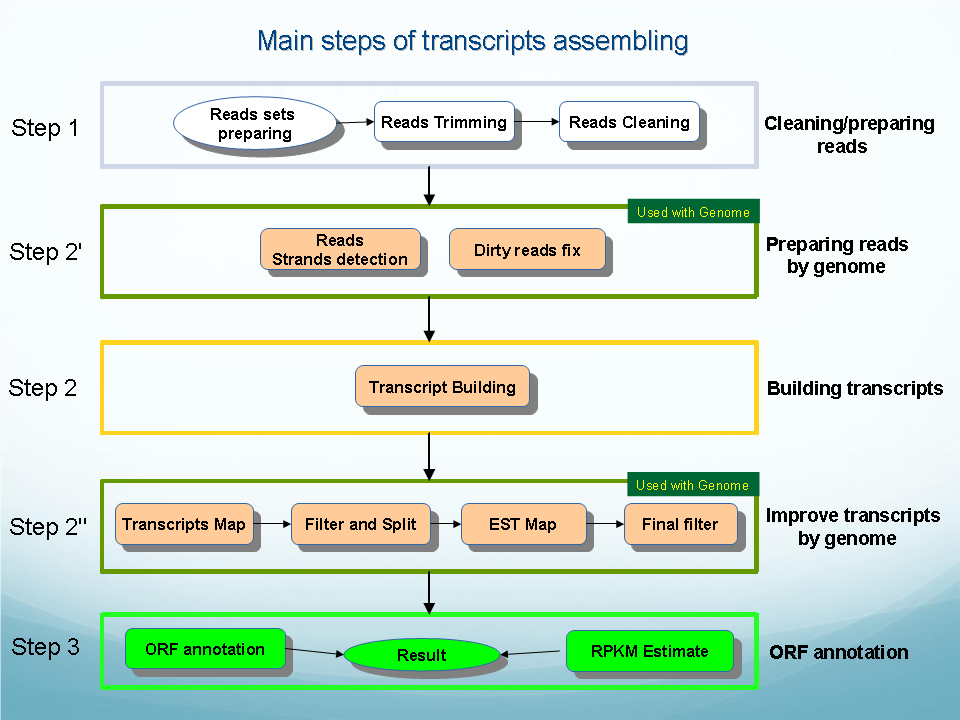
Transcripts Assembling pipeline is a tool for assembling of reads into transcripts.
Its main steps are reads trimming (trimming adapters from reads), reads cleaning (fixing reads errors) and assembling reads into transcripts (see Figure).
-
Transcripts Assembling with known genome available have the following
additional steps:
- reads strands detection (by aligning reads to genome)
- fixing reads errors (by aligning reads to genome) [in addition to fixing reads errors by aligning reads to reads]
- improving transcripts by genome
COMMANDS AND OPTIONS
Working directory is specified with an option
-work_dir:<dir>
Number of threads/processors to use is specified with an option
-j:<N>
Config files are specified with an option
-cfg:<file>
There should be usually two configuration files provided in the command line - one with default parameters for assembling such as
-cfg:reads_assembler/cfg/assembling.def.cfg
and another with user specified parameters such as
-cfg:./reads.sim.cfg
Actually, there could be any number of configuration files in the command line (all provided via -cfg:<file> option). If same parameters are defined in configuration files, parameters from next configs overwrite parameters from previous configs (as they follow in the command line).
CONFIGURATION FILES
Almost all parameters for assembling are specified in configuration files.
Configuration files have hierarchical structure.
Here is an example of 'reads.sim.cfg' with user specified parameters and
notes for them.
(For description of parameters, see also parameters of Reads Assembler.)
reads.sim.cfg
# PE reads are specified through reads.pe.<name> blocks.
# There can be as many such blocks as PE reads sets,
# for example, reads.pe.pe1, reads.pe.pe2, and so on.
# In this file there are three PE reads set 'reads.pe.pe1', 'reads.pe.pe2'
# and 'reads.pe.pe3'. These reads were simulated from transcripts of genes
# from three chromosomes - chr20, chr21, chr22.
reads.pe.pe1 {
# --trimming # trimming is not used because there were no adapters in simulated reads
--cleaning # fix reads errors, remove bad reads
--transcripts # assemble reads into transcripts
--stat-base
--stat-fastqc
src.file = "./chr20_genes_100bp_300i_40x.fa" # reads simulated from transcripts from chr20
src.format = "ifasta-pair"
# cleaning parameters for reads 'pe.pe1'
cleaning {
align_hml = "0.9" # homology threshold when aligning reads
align_err = "0.5" # error threshold (0.5 means 5 mismatches per 100 bp, non-linear parameter)
align_break = 100
align_min = 10
align_use = 10000
align_max = 100000
pos_file = "./chr20_genes_100bp_300i_40x.pos" # file for internal estimations of assembling
gen_file = "./chr20.fa" # chromosome
--best-grp
--best-clust
--clust
}
aver_pe=300
stdev_pe=30
}
reads.pe.pe2 {
# --trimming
--cleaning
--transcripts
--stat-base
--stat-fastqc
src.file = "./chr21_genes_100bp_300i_40x.fa"
src.format = "ifasta-pair"
# cleaning parameters for reads 'pe.pe2'
cleaning {
align_hml = "0.9"
align_err = "0.5"
align_break = 100
align_min = 10
align_use = 10000
align_max = 100000
pos_file = "./chr21_genes_100bp_300i_40x.pos"
gen_file = "./chr21.fa"
--best-grp
--best-clust
--clust
}
aver_pe=300
stdev_pe=30
}
reads.pe.pe3 {
# --trimming
--cleaning
--transcripts
--stat-base
--stat-fastqc
src.file = "./chr22_genes_100bp_300i_40x.fa"
src.format = "ifasta-pair"
# cleaning parameters for reads 'pe.pe3'
cleaning {
align_hml = "0.9"
align_err = "0.5"
align_break = 100
align_min = 10
align_use = 10000
align_max = 100000
pos_file = "./chr22_genes_100bp_300i_40x.pos"
gen_file = "./chr22.fa"
--best-grp
--best-clust
--clust
}
aver_pe=300
stdev_pe=30
}
hardware {
memory.max = 8 # maximal memory to use, Gb
memory.opt = 4 # optimal memory to use, Gb
}
distance {
}
# genome can be represented as one or several FASTA files (but not multi-FASTA)
genome {
src = ["chr20.fa" "chr21.fa" "chr22.fa"]
# size = 4000000
}
contigs {
# src = "./contigs.fa"
}
cleaning {
# compressing reads (--cfile) greatly speeds up calculations
# when program runs repeatedly
# (sequences represented by numbers instead of characters)
--cfile # compress reads before calculations
}
assembling {
# --cfile
--remove-overlaps
# contigs = ""
initial = "pe.pe1" # reads set to start assembling with
result = "assembling.fna" # resulting file with assembled contigs
# --ext-single-proc
# min_seq_len = 500
# remove_tail_len = 0
# remove_tail_no_paired_len = 200
--stat
--genome-match
--stat-coverage
}
scaffolding {
--stiky:off
--basic
--over
--patching
result = "scaffolding.fna"
--genome-match
--stat-coverage
}
transcripts {
--use-genome # transcripts assembling with genome
--use-unknown
# --cfile
result = "transcripts.fna" # resulting file with transcripts
min_transcript_len = 500
--stat-coverage
}
common.work.format = "fastq"
LICENSE AND CITATION
Transcripts Assembler is a free for academic usage. Please contact to softberry@softberry.com in otherwise.