Services Test Online
Analyze RNASeq Next Generation Sequencing Data:
- Accurate alignment of high-throughput RNA-seq data to a reference genome (ReadsMap);
- De novo transcriptome reads assembly into RNA transcripts (TransSeq - Program for assembling transcripts from short reads.);
Transomics - pipeline to map RNAseq data, assemble them into transcripts and quantify the abundance of these transcripts in particular datasets.
Analyze Genomic Next Generation Sequencing Data:
- OligoZip Assembler - De novo reconstruction (assembling) of genomic sequence,
- Reconstruction of sequence using reference genome,
- Mutation profiling and SNP discovery.
- Functional analysis of SNP (SNP-effect);
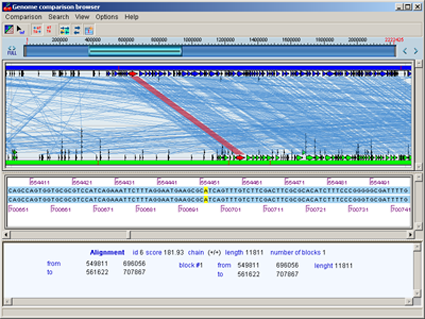
The GenomeMatch pipeline is developed to find alignments between genome sequences as well as to construct synteny maps.
GenomeMatchViewer uses output of GenomeMatch alignment program to visualize similarity of genome sequences
![]()
[Download
and Instruction] [Help] [See more pictures here]
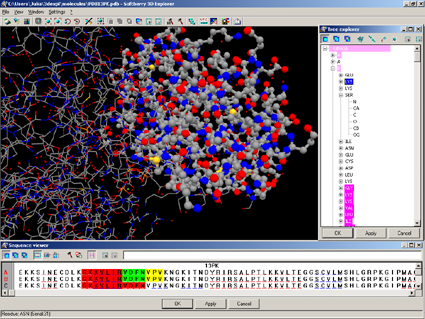
3D-Explorer is designed to visualize spatial models of biological macromolecules and their complexes. The 3D-Explorer application is compatible with PDB files. 3D-Explorer has an interface compatible with the GetAtoms and CE applications.
![]()
[Download
and Instruction] [Help] [See
more pictures here]
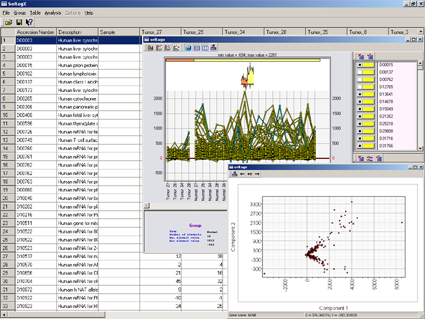
SelTag is one of the most elegant tools for analysis of expression data. It can analyze all or marked groups of genes or tissues, select tissue-specific genes based on complex criteria, provide visual representation of expression data.
GCB
[Download
and Instruction]
[Help (draft)] [See
more pictures here]
Genome Comparison Browser (GCB) is designed to visualize comparison of two genomes or chromosomes.