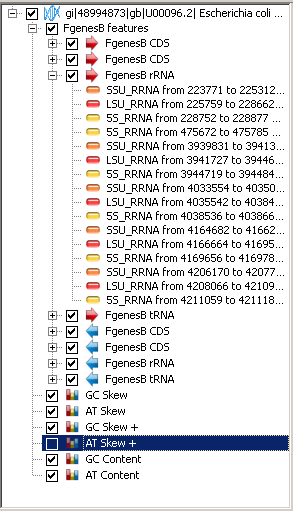
List of sequences
List of sequences contains the loaded sequences and their layout/annotation in the form of a tree (
Sequence>Feature table>Feature level>Feature) and graphical names (if any).
To set on/off the object (sequence, feature table, feature level, group etc.) representation on the map and sequence view panel,
use the ( )
checkbox near the object name. To set on/off all sequences representation use the pop-up menu commands
"Enable all" or "Disable all".
)
checkbox near the object name. To set on/off all sequences representation use the pop-up menu commands
"Enable all" or "Disable all".
Mouse double-click click on the sequence name calls out the
"Sequence properties" dialog.
Mouse right button click on the sequence
name calls out the sequence pop-up menu
with the following commands:
-
Sequence properties - call out the
"Sequence properties" dialog
for editing sequence properties/attributes.
-
Copy sequence - copy selected sequence.
-
Delete sequence - delete selected sequence.
-
New feature table - call out the "New feature table" dialog
for creating new feature table.
-
Paste feature table - paste previously copied feature table.
Feature table will be added at the end of node list.
-
Import features from GenBank - import data from Genbank file.
-
Import profile from file - add graphic data from file onto the map.
-
Add standard profiles - add standard profiles onto the map.
-
Enable all - set on all sequences representation on the map.
-
Disable all - set off all sequences representation on the map.
Mouse right button click on the feature table name calls out the feature table pop-up menu with the following commands:
-
Edit feature table - call out the
"Edit feature table" dialog
for editing feature table properties/attributes.
-
Copy feature table - copy selected feature table.
-
Delete feature table - delete selected feature table.
-
New feature level - call out the "New feature level" dialog
for creating new feature level.
Feature level will be added at the end of node list.
-
Paste feature level - paste previously copied feature level.
Feature level will be added at the end of node list.
-
Enable all - set on all sequences representation on the map.
-
Disable all - set off all sequences representation on the map.
Mouse double-click on the feature level calls out the "Edit feature level" dialog.
Mouse right button click on the feature level name calls out the feature level pop-up menu with the following commands:
-
Edit feature level - call out the
"Edit feature level" dialog
for editing feature data.
-
Copy feature level - copy selected feature level.
-
Delete feature level - delete selected feature level.
-
New feature - call out the "New feature" dialog
for creating new feature.
-
Paste feature - paste previously copied feature.
Feature will be added at the end of node list.
-
Enable all - set on all sequences representation on the map.
-
Disable all - set off all sequences representation on the map.
Mouse double-click on the feature name calls out the "Edit feature" dialog.
Mouse left button click on the feature name selects the appropriate feature on the map and on the
Sequence view panel as well as causes the information on the feature to be displayed on the Info view panel.
If the clicked feature belongs to "CDS" type, the appropriate nucleotide and aminoacid sequences are highlighted on "Sequence view panel";
if the clicked feature belongs to some other type, only nucleotide sequence is highlighted.
Mouse right button click on the feature name calls out the feature pop-up menu with the following commands:
-
Edit feature - call out the
"Edit feature" dialog
for editing feature data.
-
Copy feature - copy selected feature.
-
Delete feature - delete selected feature.
-
Fit to view - "stretch" the image of the selected feature
for all width of the map.
-
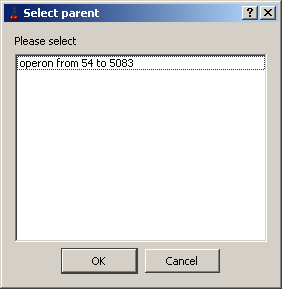
Add to parent feature -
If a feature is not a part of any other one, then in it's pop-up menu the "Add to parent feature" command appears. This command calls out
the "Select parent" dialog, where the appropriate "parent" feature can be chosen. As a parent feature, any operon that lays in the same layer
and overlaps the original feature can be used. If feature does not overlap any "operon" type one, the "No suitable parent found" message box will appear.
-
Remove from parent -
If a feature is already a part of any other one, then in it's pop-up menu the "Remove from parent" command appears.
For example, CDS that is a part of an operon has in it's pop-up menu the this command.
-
Enable all - set on all sequences representation on the map.
-
Disable all - set off all sequences representation on the map.
Mouse right button click on the profile name calls out the profile pop-up menu with the following commands:
-
Delete profile - delete selected profile.
-
Enable all - set on all sequences representation on the map.
-
Disable all - set off all sequences representation on the map.

![]() )
checkbox near the object name. To set on/off all sequences representation use the pop-up menu commands
"Enable all" or "Disable all".
)
checkbox near the object name. To set on/off all sequences representation use the pop-up menu commands
"Enable all" or "Disable all".